Projects
Application
Alzheimer's disease and robust biomarkers
Since its creation in 2011, SIMEXP has had a strong focus on clinical applications, and in particular on using rs-fMRI as a diagnostic tool for Alzheimer's disease. We first investigated the test-retest reliability of resting-state fMRI connectivity in elderly subjects at familial risk of AD (Orban et al., Scientific Data 2015) and used Monte-Carlo simulations to assess the impact of site effects on the comparison of clinical groups (Dansereau et al., Neuroimage 2017).
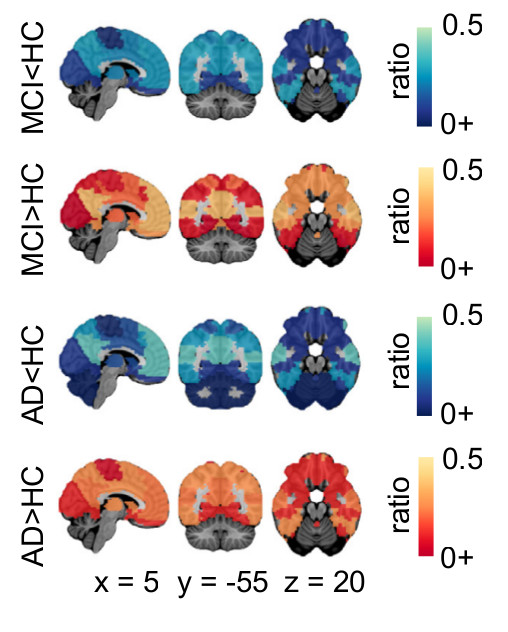
We then worked on identifying functional measures that would be good targets for AD biomarkers. We first implemented a systematic meta-analysis on cross-sectional differences between AD patients, patients with mild cognitive impairment (MCI) and cognitively normal elderly, using published literature (Badhwar et al., Alzheimer’s and dementia, 2017), as well as retrospective data pooling (Tam et al., Frontiers in Aging Neuroscience, 2015).
Application
Subtyping in Alzheimer's disease
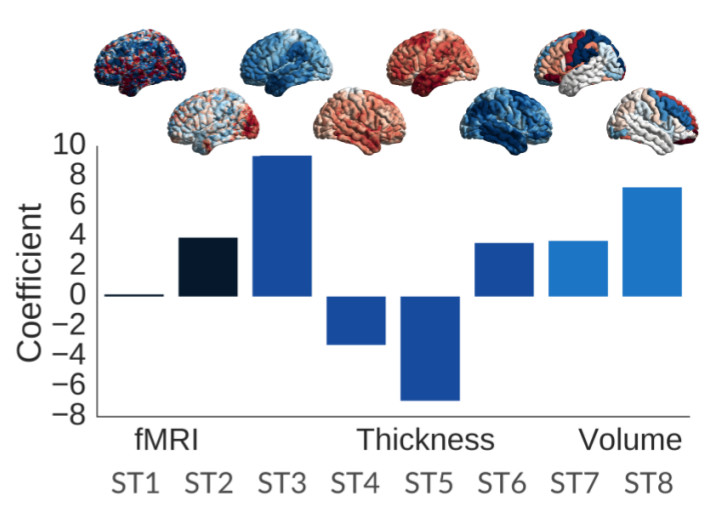
In the Tam et al. (2015) experiment, the heterogeneity in patterns of functional connectivity across sites was striking. We then looked at subtypes of functional networks and their association with clinical symptoms in patients with AD or MCI, as well as beta amyloid deposition in elderly subjects at familial risk of AD (Orban et al., biorxiv 2017). We then successfully tested the idea of identifying a subgroup of people with homogeneous multimodal brain signature, that put them highly at risk of progressing to dementia within 3 years, using machine learning models (Dansereau et al., biorxiv 2018). We validated further this idea on a large data sample, using only structural imaging and cognitive evaluation (Tam et al., 2018). As part of our work with the biomarkers team of the Canadian Consortium on Neurodegeneration in Aging (http://ccna-ccnv.ca/en/), Dr Badhwar is going to implement this approach using extensive multimodal markers on a large heterogeneous cohort recruited across Canada (N=1600).
Algorithms
Functional brain parcellation
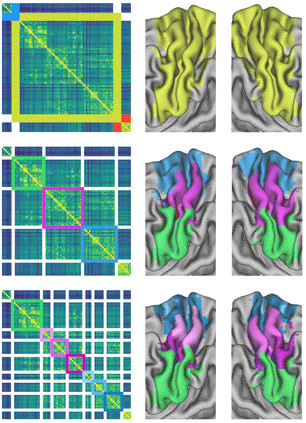
An early interest of Dr Bellec was functional brain parcellation: finding "modules" in the brain that are more strongly functionally connected within themselves than across modules. These contributions included a two-stage algorithm to work first on regions before moving to full-brain networks, as well as a bootstrap analysis of the stability of clusters (BASC), both for individual or group-level parcellations.
Since then, Tam and colleagues used BASC to generate a new parcellation template specifically adapted to older subjects, and individuals with mild cognitive impairment. Urchs and colleagues also generated the MIST functional template, which used multiple resolutions. This means that the brain is decomposed with various levels of details: from distributed networks, down to subnetworks and regions. Sebastian took great care in labelling those parcels in a consistent way across resolutions.
Finally, Amal Boukhdir's PhD work is looking to extend the BASC framework to make it more scalable, working at the voxel resolution for very long fMRI time series. She is also exploring how the brain dynamically reorganizes its modular structure over time, rather than extracting a singe individual parcellation for each subject.
Algorithms
Tasks, cognitive states and brain connectivity
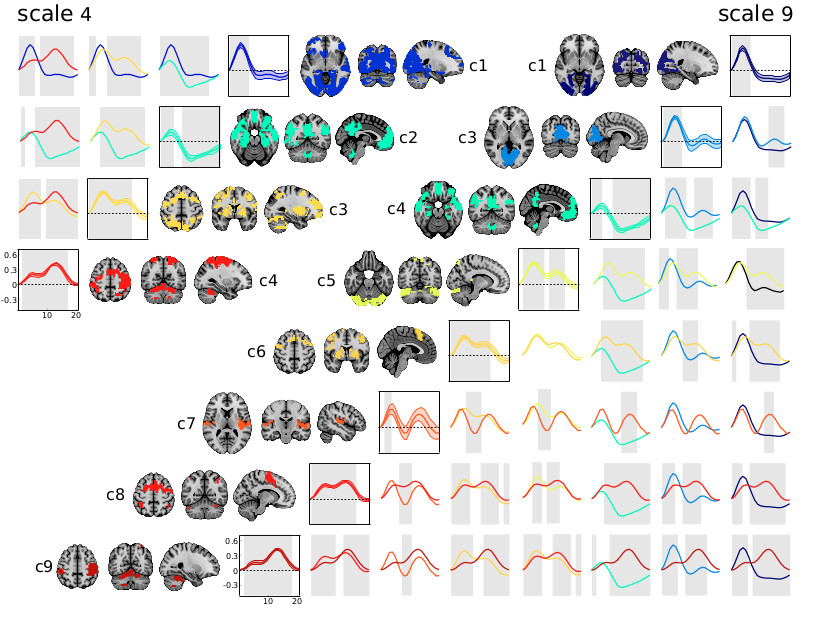
In 2015, Orban and colleagues showed that the response to a simple motor task was enough to parcellate the whole brain into a hierarchy of task-related networks. This hierarchy was largely driven by a gradient of involvement in the preparation relative to the execution of the movement. This paper demonstrated the richness of task activity captured by fMRI, and how even coarse parcellation can discriminate subtle differences in brain dynamics. As part of her post-doctoral project, Dr Yu Zhang now tries to leverage this property using deep artificial networks based on recurrent graph convolution, to predict individual variations in cognitive states from fMRI dynamics. This work is laying the grounds for the Courtois NeuroMod project, which will use extensive fMRI and MEG data to help train large recurrent artificial neural networks on a variety of tasks.
Open Science
Code, Data and Idea Sharing
The laboratory is very committed to open science principles, being a co-founder of the open science special interest group at the Organization for Human Brain Mapping, and a member of the Canadian Open Neuroscience Platform. We are sharing free software: the Neuroimaging Analysis Kit (NIAK), for preprocessing and connectome analysis of fMRI data; and the Pipeline System for Octave and Matlab (PSOM)), for high-throughput parallel analysis. We are also sharing raw data, e.g. a test-retest dataset with older subjects as part of the CoRR consortium, Orban et al. 2015 ; preprocessed data, e.g. ADHD200 preprocessed; as well as data derivatives such as multiresolution parcellations (Urchs et al. 2018 ). Finally, the laboratory is actively participating in, and/or organizing, many open science events, notably brainhack and the brainhack school.
